Molecular Biology | Bioinformatics
Molecular Taxonomy of Mirids

Background
While humans are equipped with the intelligence to make sophisticated machines and travel into space, insects really rule the world. With an estimated 10 million species and more biomass than humans, insects are the most successful creatures on Earth. While the reason for their success remains an important subject of scientific research, there is still an enormous gap in our understanding of insect taxonomy (the identification and description of species and other taxa). Insects in the family Miridae are known as plant bugs, and are part of a large group of insects called “true bugs” (suborder Heteroptera) that feed by sucking plant (and sometimes animal) juices through their mouth, a straw-like tube. Plant bugs represent the largest Heteropteran family with over 11,000 described species in about 1,400 genera (Wheeler, 2001). While the oldest Homo sapiens fossils date back to about 195,000 years ago (McDougall et al., 2005), plant bugs have roamed the Earth since the Jurassic period 199-145 million years ago (Wheeler, 2001). This study focuses on a group of plant bugs in the genus Pseudoloxops, which are diverse, understudied and endemic to the island of Tahiti, French Polynesia. Tahiti is a 403 mi2 volcanic island located in the Archipelago of the Society Islands in the southern Pacific Ocean, and harbors more endemic (i.e. found nowhere else) insect species than any other island in French Polynesia. Five species of Pseudoloxops were previously known from Tahiti (Knight, 1937), and were described based on morphological data alone, especially color patterns. Here we take an integrative taxonomic approach by combining morphological, molecular and ecological data as lines of evidence to delimit species (Gebiola et al., 2011). We use molecular genetic data as our primary line of evidence, and follow the genealogical species concept, which groups organisms into species based on their common ancestry (Shaw, 2001). The main goal of this project is to find out how many species of Pseudoloxops there are on the island of Tahiti.
Methods
- We collected 18 adult specimens of Pseudoloxops representing a wide range of morphological variation in 2008, 2009, and 2011 from the island of Tahiti. We also identified the host plant that each specimen was collected from.
- Specimens were given a unique identifier (e.g. Z114) and stored in 95% alcohol in a -80°C freezer to preserve DNA.
- We extracted DNA and amplified it using the Polymerase Chain Reaction (PCR) for segments of two genes: 814 base pairs of COI (cytochrome C Oxidase I), a mitochondrial gene; and 637 base pairs of 28S, a nuclear ribosomal gene.
- We ran gel electrophoresis on the PCR products to verify DNA amplification.
- 18 successful PCR products were sent to the UC Berkeley DNA Sequencing Facility for sequencing.
- Specimen sequences were aligned in the program Geneious Pro 5.6.2 .
- The aligned sequences for each gene were entered into Geneious’ MrBayes plug-in to build phylogenetic trees based on Bayesian inference (Huelsenbeck and Ronquist, 2001).
- The two sequence alignments were combined to produce a single 50% majority rules consensus phylogeny for all 18 specimens, plus 1 outgroup specimen (Z16). Following the genealogical species concept, we considered a species to be any well-supported (i.e. posterior probability ≥80%) monophyletic clade that was ≥1% genetically distinct from its sister clade.
- After identifying putative species, we examined the color patterns and host-plant associations of the individuals within each species to see if morphology and ecology matched with the molecular data.
Results and Discussion
We successfully amplified 17 of 18 specimens for each gene; 135 of 814 (16.6%) base pairs for CO1 were variable; 18 of 637 (2.8%) base pairs were variable for 28S. Gene sequences that were not amplified were coded as missing data. The phylogeny showed 5 distinct, well-supported monophyletic groups that we propose correspond to 5 species. (figure below)
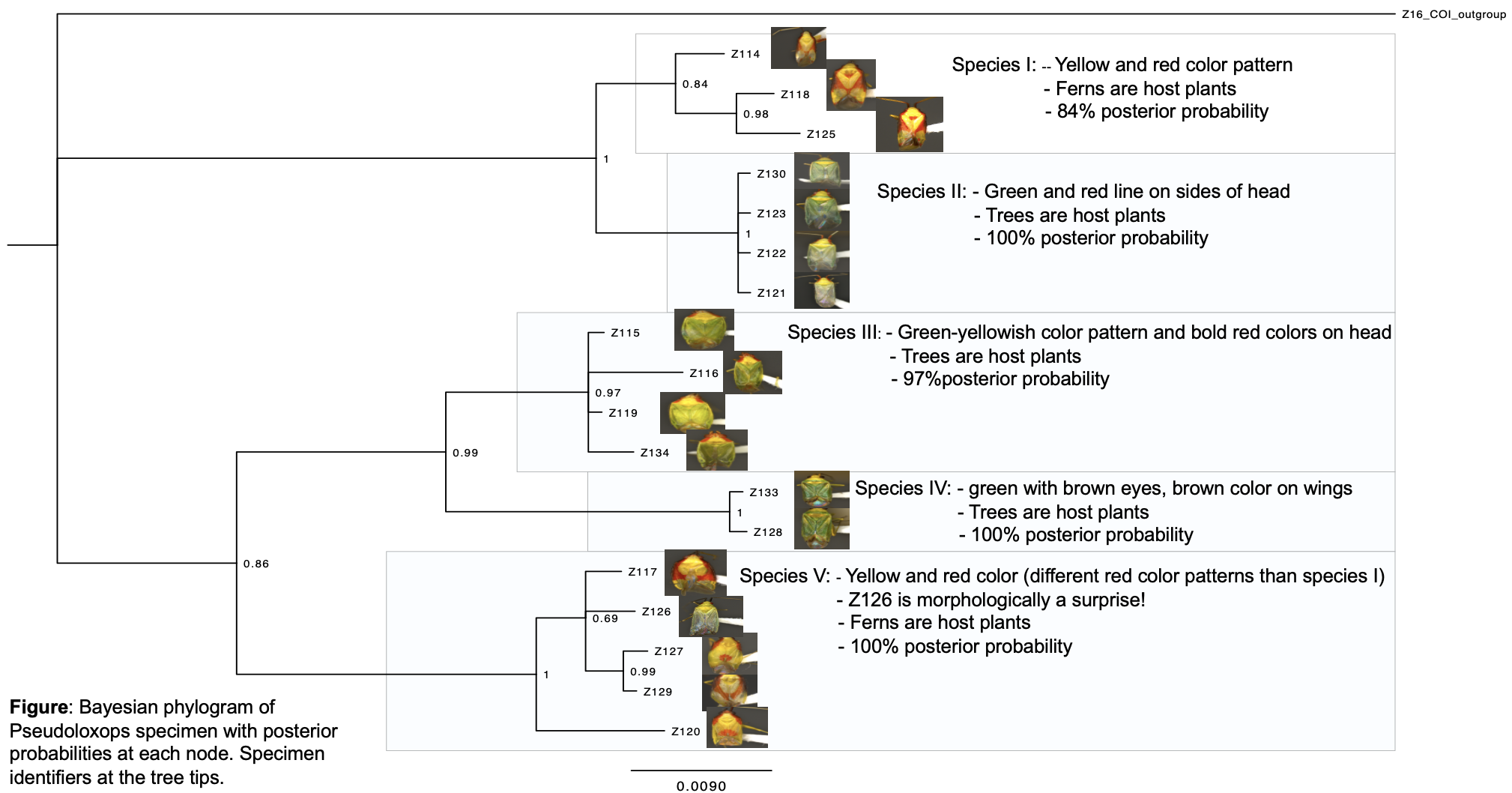
- While one of the proposed new species looks morphologically similar to the previously described species, the other four species look distinctly different, suggesting that there are at least four species new to science awaiting formal description.
- Considering that the scope of the study was limited to Tahiti, collections from neighboring islands may reveal further new species that are closely related to those included in this study.
- The difference in coloration between species associated with trees and those associated with ferns suggests that the speciation process of these plant bugs may have been driven by natural selection; color could be used as camouflage to hide from predators.
- Our small sample size does not provide enough evidence to make a final determination of the number of Pseudoloxops species currently living in Tahiti. Adding the additional components of this larger integrative taxonomy study will provide conclusive results on the evolution and speciation of Tahitian plant bugs.